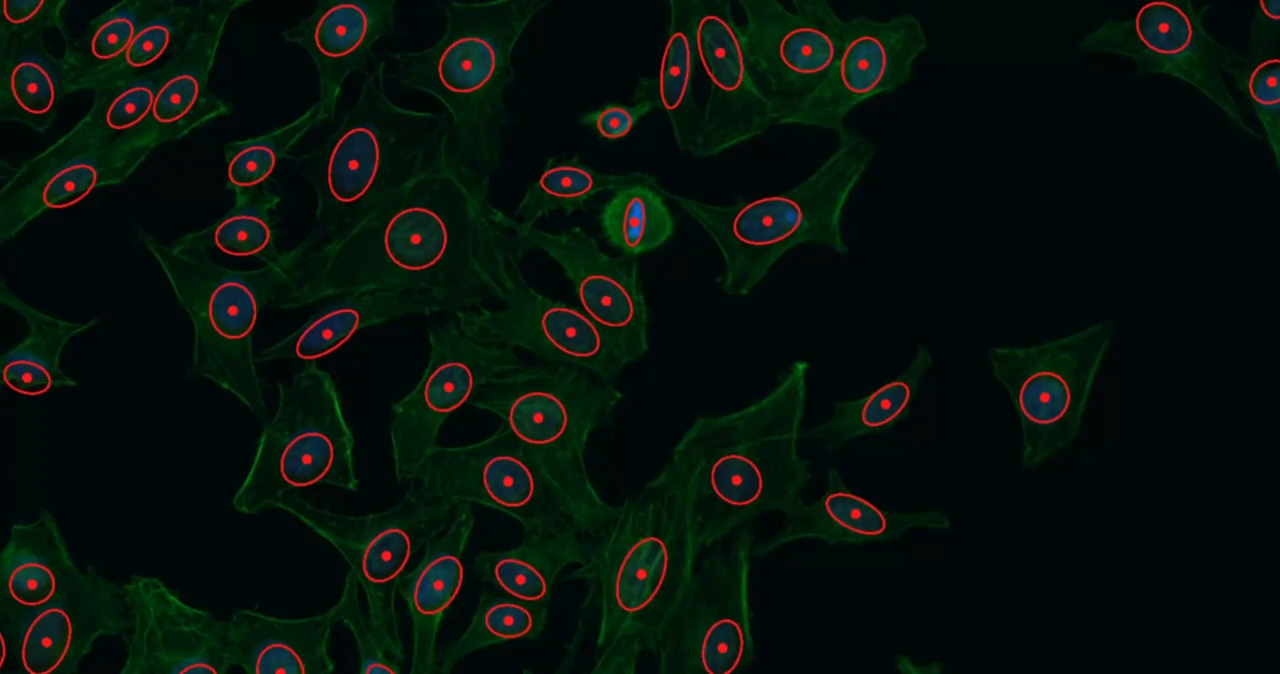
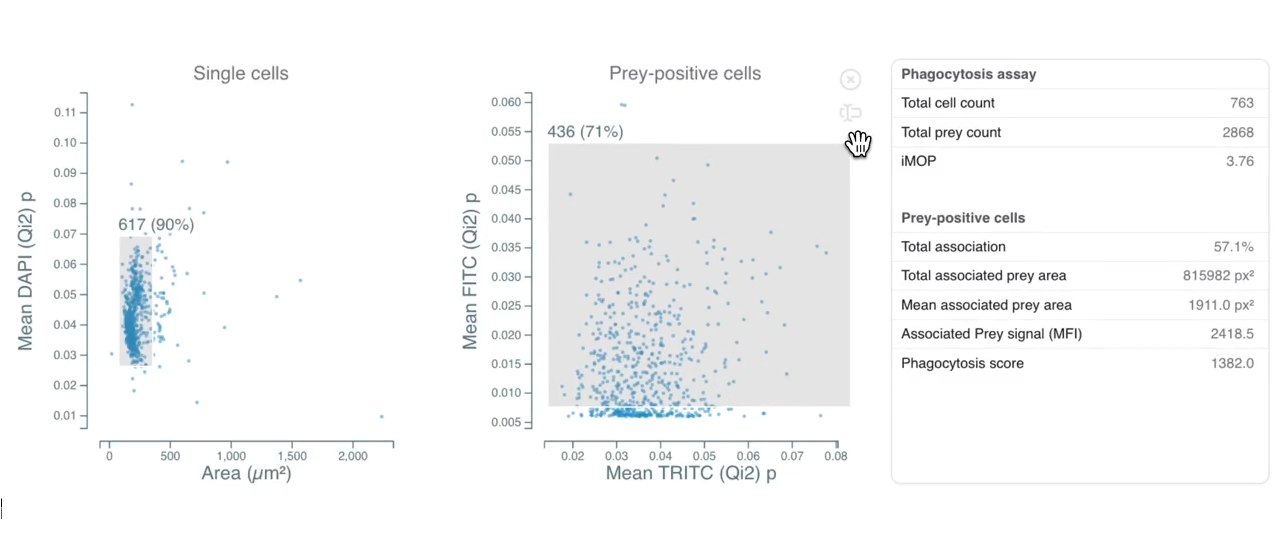
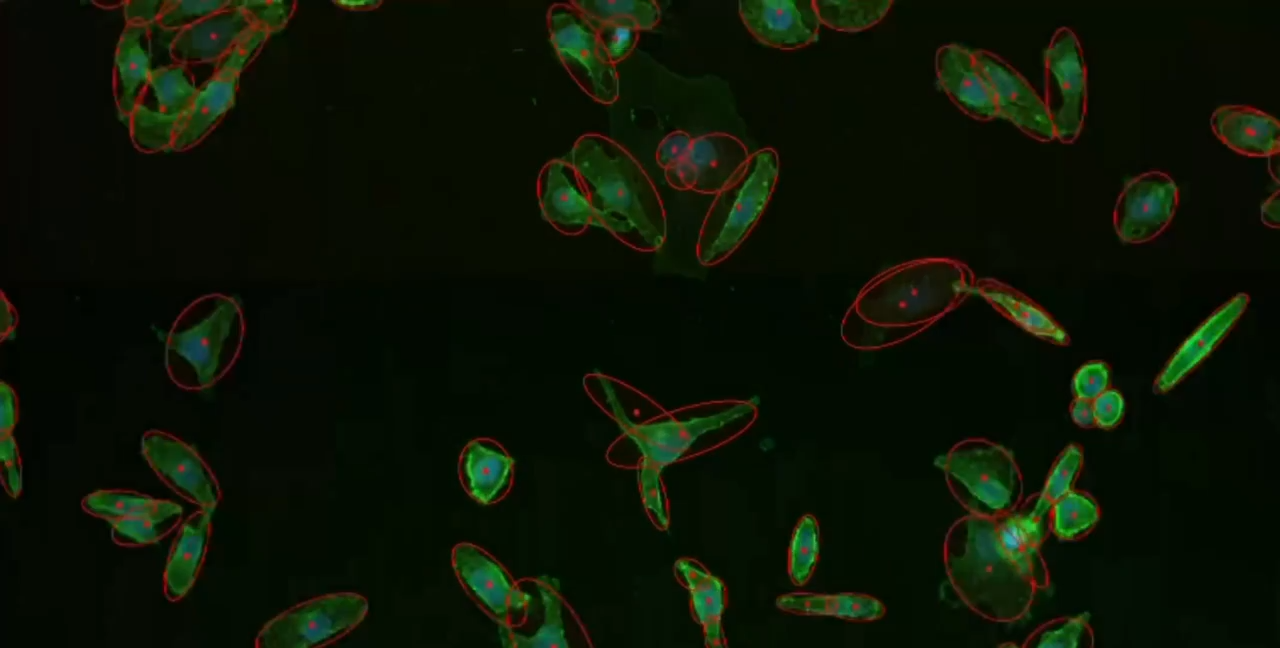
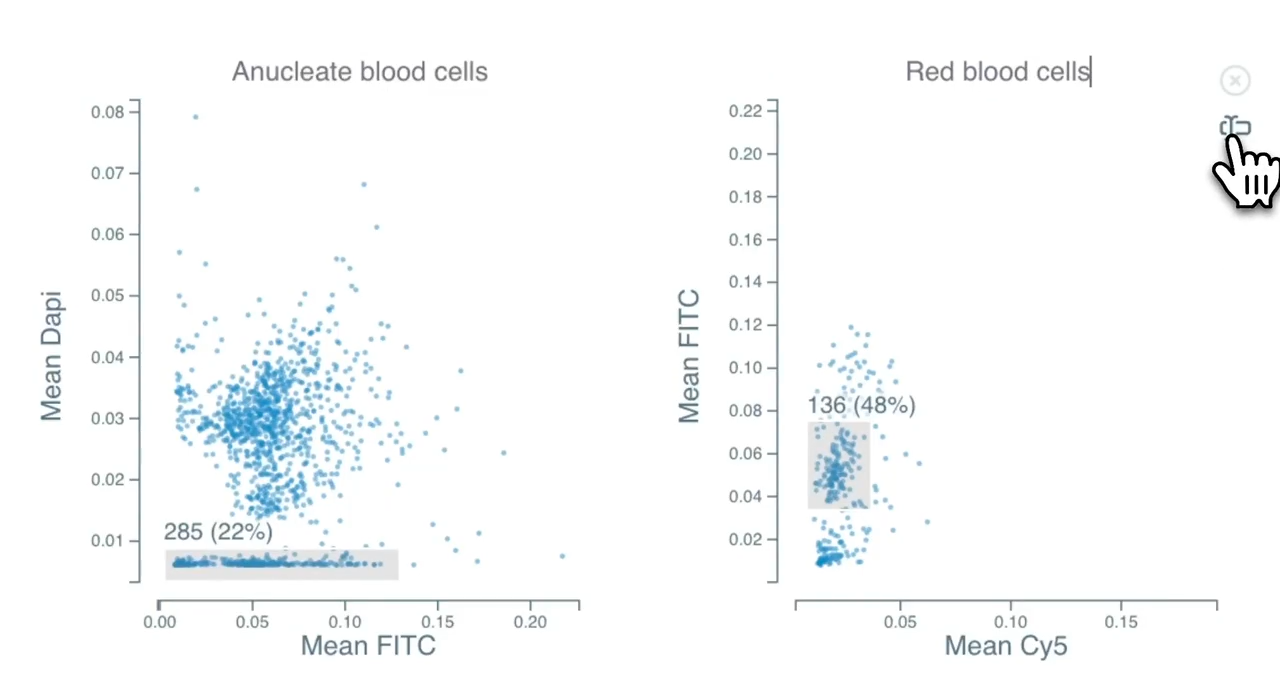
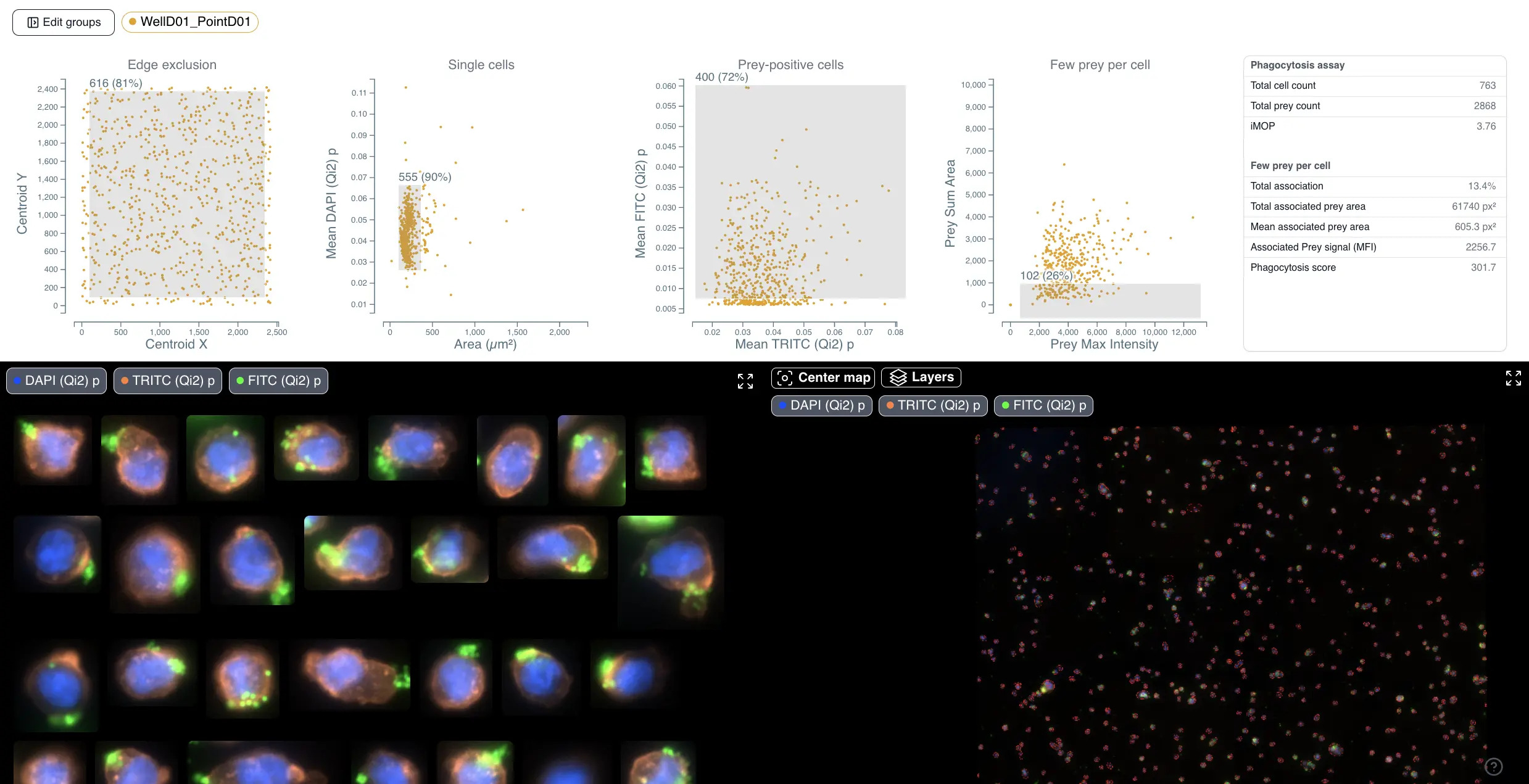
Phagocytosis is a fundamental cellular process crucial for immune defense, tissue homeostasis, development, and diseases like cancer. The process involves several steps: recognition of the target particle via receptors (e.g., Fc receptors, complement receptors, scavenger receptors), signaling to activate the internalization machinery, formation of a phagosome through actin-driven protrusions, and finally maturation into a phagolysosome for degradation.
Despite its importance, quantifying phagocytosis is notoriously challenging with traditional methods. Manual counts are time-consuming, subjective, and suffer from poor reproducibility. Flow cytometry, while offering high throughput, can yield artifacts, struggles to differentiate between bound and internalized particles, and rarely provides kinetic information. Many methods provide only static snapshots, missing the dynamic nature of the process. Distinguishing between association (binding) and actual internalization is often difficult. Furthermore, analysis software like ImageJ or CellProfiler often requires significant expertise and time to configure and validate robust analysis pipelines, further complicating reproducible quantification.
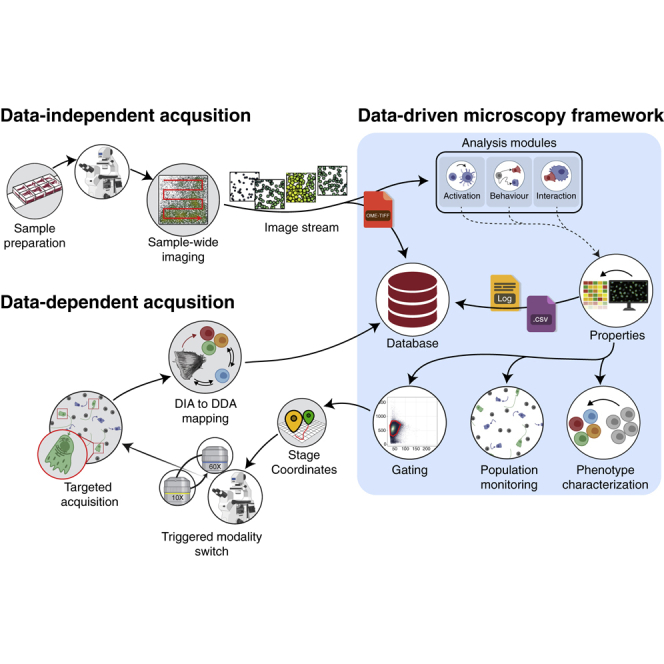
Cytely addresses these challenges by implementing a fully automated Smart Microscopy workflow. This approach combines intelligent image acquisition (separating initial scans from targeted follow-up) with sophisticated real-time analysis to deliver robust, quantitative, and dynamic information about phagocytosis with minimal user intervention.